Log Link vs Log Transformation in R — The Difference that Misleads Your Entire Data Analysis
Although normal distributions are the most commonly used, a lot of real-world data unfortunately is not normal. When faced with extremely skewed data, it’s tempting for us to utilize log transformations to normalize the distribution and stabilize the variance. I recently worked on a project analyzing the energy consumption of training AI models, using data […] The post Log Link vs Log Transformation in R — The Difference that Misleads Your Entire Data Analysis appeared first on Towards Data Science.


To address this skewness and heteroskedasticity, my first instinct was to apply a log transformation to the Energy variable. The distribution of log(Energy) looked much more normal (Fig. 2), and a Shapiro-Wilk test confirmed the borderline normality (p ≈ 0.5).

Modeling Dilemma: Log Transformation vs Log Link
The visualization looked good, but when I moved on to modeling, I faced a dilemma: Should I model the log-transformed response variable (log(Y) ~ X), or should I model the original response variable using a log link function (Y ~ X, link = “log")? I also considered two distributions — Gaussian (normal) and Gamma distributions — and combined each distribution with both log approaches. This gave me four different models as below, all fitted using R’s Generalized Linear Models (GLM):
all_gaussian_log_link <- glm(Energy_kWh ~ Parameters +
Training_compute_FLOP +
Training_dataset_size +
Training_time_hour +
Hardware_quantity +
Training_hardware,
family = gaussian(link = "log"), data = df)
all_gaussian_log_transform <- glm(log(Energy_kWh) ~ Parameters +
Training_compute_FLOP +
Training_dataset_size +
Training_time_hour +
Hardware_quantity +
Training_hardware,
data = df)
all_gamma_log_link <- glm(Energy_kWh ~ Parameters +
Training_compute_FLOP +
Training_dataset_size +
Training_time_hour +
Hardware_quantity +
Training_hardware + 0,
family = Gamma(link = "log"), data = df)
all_gamma_log_transform <- glm(log(Energy_kWh) ~ Parameters +
Training_compute_FLOP +
Training_dataset_size +
Training_time_hour +
Hardware_quantity +
Training_hardware + 0,
family = Gamma(), data = df)Model Comparison: AIC and Diagnostic Plots
I compared the four models using Akaike Information Criterion (AIC), which is an estimator of prediction error. Typically, the lower the AIC, the better the model fits.
AIC(all_gaussian_log_link, all_gaussian_log_transform, all_gamma_log_link, all_gamma_log_transform)
df AIC
all_gaussian_log_link 25 2005.8263
all_gaussian_log_transform 25 311.5963
all_gamma_log_link 25 1780.8524
all_gamma_log_transform 25 352.5450Among the four models, models using log-transformed outcomes have much lower AIC values than the ones using log links. Since the difference in AIC between log-transformed and log-link models was substantial (311 and 352 vs 1780 and 2005), I also examined the diagnostics plots to further validate that log-transformed models fit better:




Based on the AIC values and diagnostic plots, I decided to move forward with the log-transformed Gamma model, as it had the second-lowest AIC value and its Residuals vs Fitted plot looks better than that of the log-transformed Gaussian model.
I proceeded to explore which explanatory variables were useful and which interactions may have been significant. The final model I selected was:
glm(formula = log(Energy_kWh) ~ Training_time_hour * Hardware_quantity +
Training_hardware + 0, family = Gamma(), data = df)Interpreting Coefficients
However, when I started interpreting the model’s coefficients, something felt off. Since only the response variable was log-transformed, the effects of the predictors are multiplicative, and we need to exponentiate the coefficients to convert them back to the original scale. A one-unit increase in





































































![New iPad 11 (A16) On Sale for Just $277.78! [Lowest Price Ever]](https://www.iclarified.com/images/news/97273/97273/97273-640.jpg)

![Apple Foldable iPhone to Feature New Display Tech, 19% Thinner Panel [Rumor]](https://www.iclarified.com/images/news/97271/97271/97271-640.jpg)


























-xl.jpg)





























































































































































































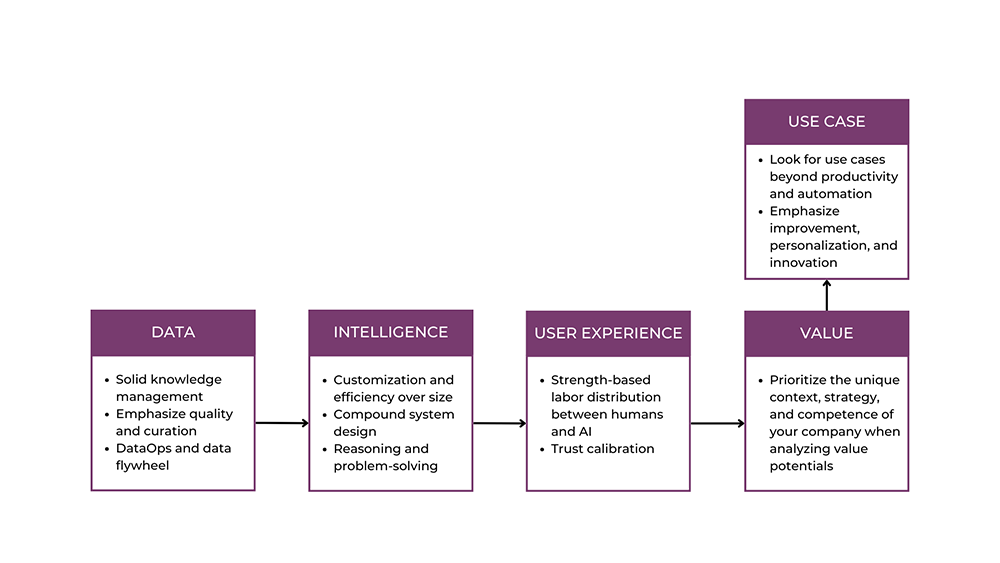

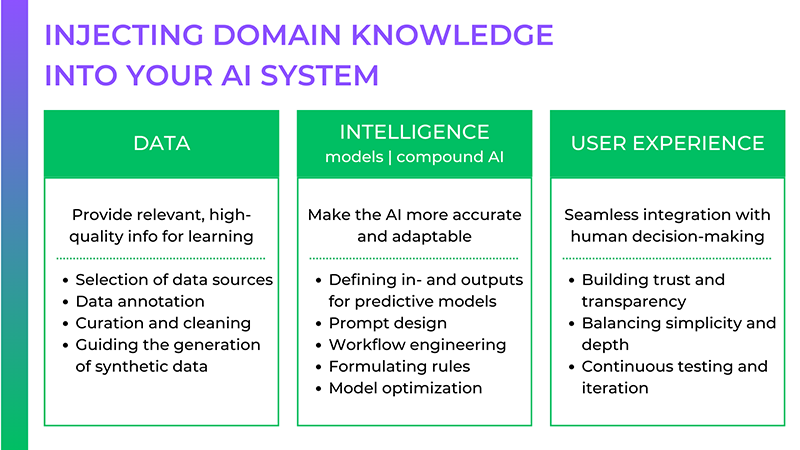



![[The AI Show Episode 146]: Rise of “AI-First” Companies, AI Job Disruption, GPT-4o Update Gets Rolled Back, How Big Consulting Firms Use AI, and Meta AI App](https://www.marketingaiinstitute.com/hubfs/ep%20146%20cover.png)






























































































































![Ditching a Microsoft Job to Enter Startup Hell with Lonewolf Engineer Sam Crombie [Podcast #171]](https://cdn.hashnode.com/res/hashnode/image/upload/v1746753508177/0cd57f66-fdb0-4972-b285-1443a7db39fc.png?#)



























-Nintendo-Switch-2-Hands-On-Preview-Mario-Kart-World-Impressions-&-More!-00-10-30.png?width=1920&height=1920&fit=bounds&quality=70&format=jpg&auto=webp#)


















































.jpg?width=1920&height=1920&fit=bounds&quality=70&format=jpg&auto=webp#)















