Medical RAG Research with txtai
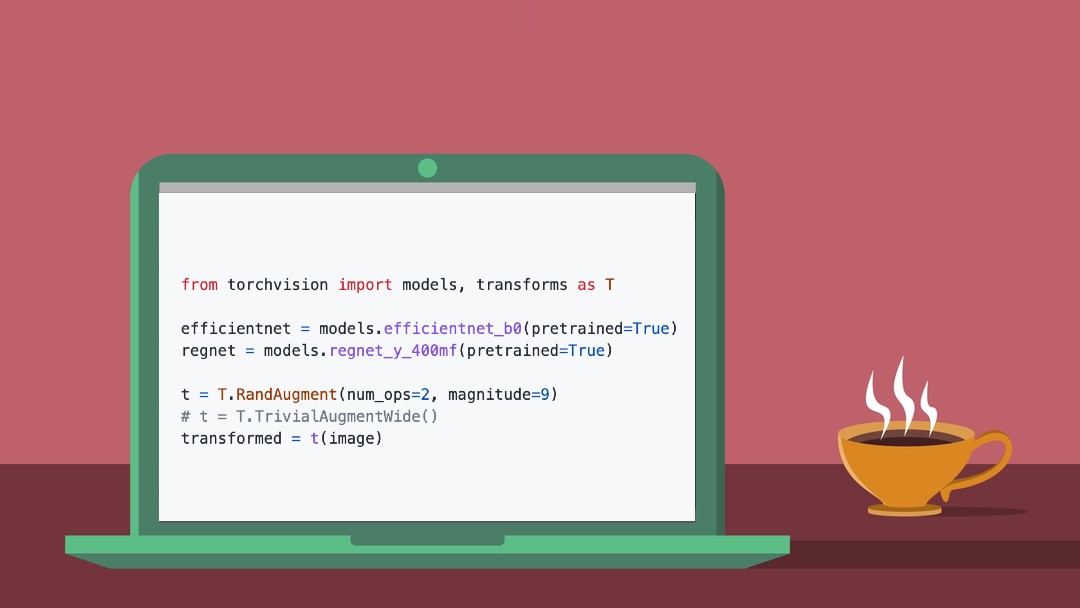
txtai is an all-in-one AI framework for semantic search, LLM orchestration and language model workflows. Large Language Models (LLMs) have captured the public's attention with their impressive capabilities. The Generative AI era has reached a fever pitch with some predicting the coming rise of superintelligence. LLMs are far from perfect though and we're still a ways away from true AI. One big challenge is with hallucinations. Hallucinations is the term for when an LLM generates output that is factually incorrect. The alarming part of this is that on a cursory glance, it actually sounds like factual content. The default behavior of LLMs is to produce plausible answers even when no plausible answer exists. LLMs are not great at saying I don't know. Retrieval Augmented Generation (RAG) helps reduce the risk of hallucinations by limiting the context in which a LLM can generate answers. This is typically done with a search query that hydrates a prompt with a relevant context. RAG has been one of the most practical use cases of the Generative AI era. This article will demonstrate how to build a Medical RAG Research process with txtai. Install dependencies Install txtai and all dependencies. pip install txtai Medical Dataset For this example, we'll use a PubMed subset of article metadata for H5N1. This dataset was created using paperetl, an open-source library for parsing medical and scientific papers. PubMed has over 38 million article abstracts as of June 2025. paperetl supports loading the full dataset with all 38 million articles or just a smaller subset. The dataset link above has more details on how this can be changed for different codes and keywords. This link also has information on how the article abstracts can be loaded in addition to the metadata. from datasets import load_dataset from txtai import Embeddings ds = load_dataset("neuml/pubmed-h5n1", split="train") Next, we'll build a txtai embeddings index with the articles. We'll use a vector embeddings model that specializes in vectorizing medical papers: PubMedBERT Embeddings. embeddings = Embeddings(path="neuml/pubmedbert-base-embeddings", content=True, columns={"text": "title"}) embeddings.index(x for x in ds if x["title"]) embeddings.count() 7865 RAG Pipeline There are a number of prior examples on how to run RAG with txtai. The RAG pipeline takes two main parameters, an embeddings database and an LLM. The embeddings database is the one just created above. For this example, we'll use a simple local LLM with 600M parameters. Substitute your own embeddings database to change the knowledge base. txtai supports running local LLMs via transformers or llama.cpp. It also supports a wide variety of LLMs via LiteLLM. For example, setting the 2nd RAG pipeline parameter below to gpt-4o along with the appropriate environment variables with access keys switches to a hosted LLM. See this documentation page for more on this. from txtai import RAG # Prompt templates system = "You are a friendly medical assistant that answers questions" template = """ Answer the following question using the provided context. Question: {question} Context: {context} """ # Create RAG pipeline rag = RAG(embeddings, "Qwen/Qwen3-0.6B", system=system, template=template, output="flatten") RAG Queries Now that the pipeline is setup, let's run a query. print(rag("Tell me about H5N1")) Okay, let's see. The user is asking about H5N1. The context provided starts with "Why tell me now?" and then goes into facts about H5N1. The first sentence mentions that people and healthcare providers are weighing in on pandemic messages. Then it says H5N1 is avian influenza, a potential pandemic. Wait, but the user's question is about H5N1. The context doesn't go into specifics about what H5N1 is, but it does state that it's avian influenza. So I need to make sure I answer based on that. The answer should be concise, maybe mention that H5N1 is avian flu and it's a potential pandemic. Also, note that people are weighing in on messages. But I need to check if there's any more information. The context ends there. So the answer should be straightforward. H5N1 influenza viruses are a type of avian influenza, a potential pandemic influenza virus that could cause widespread illness and death. While the context highlights the importance of public health and preparedness, it does not provide more specific details about its characteristics or risks. Notice that this LLM outputs a thinking or reasoning section then the answer. Let's review the context to validate this answer is derived from the knowledge base. embeddings.search("Tell me about H5N1", limit=10) [{'id': '16775537', 'text': '"Why tell me now?" the public and healthcare providers weigh in on pandemic influenza messages.', 'score': 0.7156285643577576}, {'id': '22308474', 'text': 'H5N1 influenza viruses: facts

txtai is an all-in-one AI framework for semantic search, LLM orchestration and language model workflows.
Large Language Models (LLMs) have captured the public's attention with their impressive capabilities. The Generative AI era has reached a fever pitch with some predicting the coming rise of superintelligence.
LLMs are far from perfect though and we're still a ways away from true AI. One big challenge is with hallucinations. Hallucinations is the term for when an LLM generates output that is factually incorrect. The alarming part of this is that on a cursory glance, it actually sounds like factual content. The default behavior of LLMs is to produce plausible answers even when no plausible answer exists. LLMs are not great at saying I don't know.
Retrieval Augmented Generation (RAG) helps reduce the risk of hallucinations by limiting the context in which a LLM can generate answers. This is typically done with a search query that hydrates a prompt with a relevant context. RAG has been one of the most practical use cases of the Generative AI era.
This article will demonstrate how to build a Medical RAG Research process with txtai.
Install dependencies
Install txtai and all dependencies.
pip install txtai
Medical Dataset
For this example, we'll use a PubMed subset of article metadata for H5N1. This dataset was created using paperetl, an open-source library for parsing medical and scientific papers.
PubMed has over 38 million article abstracts as of June 2025. paperetl supports loading the full dataset with all 38 million articles or just a smaller subset. The dataset link above has more details on how this can be changed for different codes and keywords. This link also has information on how the article abstracts can be loaded in addition to the metadata.
from datasets import load_dataset
from txtai import Embeddings
ds = load_dataset("neuml/pubmed-h5n1", split="train")
Next, we'll build a txtai embeddings index with the articles. We'll use a vector embeddings model that specializes in vectorizing medical papers: PubMedBERT Embeddings.
embeddings = Embeddings(path="neuml/pubmedbert-base-embeddings", content=True, columns={"text": "title"})
embeddings.index(x for x in ds if x["title"])
embeddings.count()
7865
RAG Pipeline
There are a number of prior examples on how to run RAG with txtai. The RAG pipeline takes two main parameters, an embeddings database and an LLM. The embeddings database is the one just created above. For this example, we'll use a simple local LLM with 600M parameters.
Substitute your own embeddings database to change the knowledge base. txtai supports running local LLMs via transformers or llama.cpp. It also supports a wide variety of LLMs via LiteLLM. For example, setting the 2nd RAG pipeline parameter below to gpt-4o along with the appropriate environment variables with access keys switches to a hosted LLM. See this documentation page for more on this.
from txtai import RAG
# Prompt templates
system = "You are a friendly medical assistant that answers questions"
template = """
Answer the following question using the provided context.
Question:
{question}
Context:
{context}
"""
# Create RAG pipeline
rag = RAG(embeddings, "Qwen/Qwen3-0.6B", system=system, template=template, output="flatten")
RAG Queries
Now that the pipeline is setup, let's run a query.
print(rag("Tell me about H5N1"))
Okay, let's see. The user is asking about H5N1. The context provided starts with "Why tell me now?" and then goes into facts about H5N1. The first sentence mentions that people and healthcare providers are weighing in on pandemic messages. Then it says H5N1 is avian influenza, a potential pandemic.
Wait, but the user's question is about H5N1. The context doesn't go into specifics about what H5N1 is, but it does state that it's avian influenza. So I need to make sure I answer based on that. The answer should be concise, maybe mention that H5N1 is avian flu and it's a potential pandemic. Also, note that people are weighing in on messages. But I need to check if there's any more information. The context ends there. So the answer should be straightforward.
H5N1 influenza viruses are a type of avian influenza, a potential pandemic influenza virus that could cause widespread illness and death. While the context highlights the importance of public health and preparedness, it does not provide more specific details about its characteristics or risks.
Notice that this LLM outputs a thinking or reasoning section then the answer.
Let's review the context to validate this answer is derived from the knowledge base.
embeddings.search("Tell me about H5N1", limit=10)
[{'id': '16775537',
'text': '"Why tell me now?" the public and healthcare providers weigh in on pandemic influenza messages.',
'score': 0.7156285643577576},
{'id': '22308474',
'text': 'H5N1 influenza viruses: facts, not fear.',
'score': 0.658343493938446},
{'id': '16440117',
'text': 'Avian influenza--a pandemic waiting to happen?',
'score': 0.5827972888946533},
{'id': '20667302',
'text': 'The influenza A(H5N1) epidemic at six and a half years: 500 notified human cases and more to come.',
'score': 0.5593500137329102},
{'id': '18936262',
'text': 'What Australians know and believe about bird flu: results of a population telephone survey.',
'score': 0.5568690299987793},
{'id': '30349811',
'text': 'Back to the Future: Lessons Learned From the 1918 Influenza Pandemic.',
'score': 0.5540266036987305},
{'id': '17276785',
'text': 'Pandemic influenza: what infection control professionals should know.',
'score': 0.5519200563430786},
{'id': '16681227',
'text': 'A pandemic flu: not if, but when. SARS was the wake-up call we slept through.',
'score': 0.5518345832824707},
{'id': '22402712',
'text': 'Ferretting out the facts behind the H5N1 controversy.',
'score': 0.5508109331130981},
{'id': '25546511',
'text': "One-way trip: influenza virus' adaptation to gallinaceous poultry may limit its pandemic potential.",
'score': 0.5494509339332581}]
The answer is doing a good job being based on the context above. Also keep in mind this is a small 600M parameter model, which is even more impressive.
Let's try another query.
print(rag("What locations have had H5N1 outbreaks?"))
Okay, let's see. The user is asking about the locations that have had H5N1 outbreaks, and the provided context mentions a few places: Indonesia and Bangladesh. The context also has a title about a decade of avian influenza in Bangladesh and mentions "H5N1."
Wait, the user's question is in English, so I need to make sure I'm interpreting the context correctly. The context includes two sentences: one about a decade in Bangladesh and another about H5N1. The user is probably looking for specific locations where H5N1 has been reported.
Looking at the context again, it says "Human avian influenza in Indonesia" and "A Decade of Avian Influenza in Bangladesh: Where Are We Now? Are we ready for pandemic influenza H5N1?" So the outbreaks are in Indonesia and Bangladesh.
I should confirm that there are no other mentions of other locations. The context doesn't provide more information beyond those two countries. Therefore, the answer should list Indonesia and Bangladesh as the locations with H5N1 outbreaks.
The locations with H5N1 outbreaks are Indonesia and Bangladesh.
embeddings.search("What locations have had H5N1 outbreaks?", limit=10)
[{'id': '21706937',
'text': 'Human avian influenza in Indonesia: are they really clustered?',
'score': 0.6269429326057434},
{'id': '31514405',
'text': 'A Decade of Avian Influenza in Bangladesh: Where Are We Now?',
'score': 0.5972536206245422},
{'id': '15889987',
'text': 'Are we ready for pandemic influenza H5N1?',
'score': 0.5863772630691528},
{'id': '17717543',
'text': 'Commentary: From scarcity to abundance: pandemic vaccines and other agents for "have not" countries.',
'score': 0.5844159126281738},
{'id': '22491771',
'text': 'Two years after pandemic influenza A/2009/H1N1: what have we learned?',
'score': 0.5812581777572632},
{'id': '39666804',
'text': "Why hasn't the bird flu pandemic started?",
'score': 0.5738048553466797},
{'id': '23402131',
'text': 'Where do avian influenza viruses meet in the Americas?',
'score': 0.5638074278831482},
{'id': '20667302',
'text': 'The influenza A(H5N1) epidemic at six and a half years: 500 notified human cases and more to come.',
'score': 0.560465395450592},
{'id': '17338983',
'text': 'Human avian influenza: how ready are we?',
'score': 0.555113673210144},
{'id': '24518630',
'text': 'Recognizing true H5N1 infections in humans during confirmed outbreaks.',
'score': 0.5501888990402222}]
Once again the answer is based on the context which mentions the two countries in the answer. The context also discusses the Americas but it doesn't have as strong of language connecting H5N1 outbreaks to the location.
Add citations
The last item we'll cover is citations. One of the most important aspects of a RAG process is being able to ensure the answer is based on reality. There are a number of ways to do this but in this example, we'll ask the LLM to perform this step.
# Prompt templates
system = "You are a friendly medical assistant that answers questions"
template = """
Answer the following question using the provided context.
After the answer, write a citation section with ALL the original article ids used for the answer.
Question:
{question}
Context:
{context}
"""
def context(question):
context = []
for x in embeddings.search(question, limit=10):
context.append(f"ARTICLE ID: {x['id']}, TEXT: {x['text']}")
return context
# Create RAG pipeline
rag = RAG(embeddings, "Qwen/Qwen3-0.6B", system=system, template=template, output="flatten")
question = "What is H5N1?"
print(rag(question, context(question), maxlength=2048, stripthink=True))
H5N1 is a type of avian influenza virus.
**Citation Section:**
- ARTICLE ID: 22010536, TEXT: Is avian influenza virus A(H5N1) a real threat to human health?
As expected, the answer adds a citation section. Also note that the RAG pipeline stripped the thinking section from the result.
Wrapping up
This article covered how to build a Medical RAG Research process with txtai. It also covered how to modify this logic to add in your own knowledge base or use a more sophisticated LLM.
With an important space such as the medical domain, it's vital to ensure that answers are derived from reliable knowledge. This article shows how to add that reliability via RAG. But as with anything in an important domain, there should be a human in the loop and answers shouldn't be blindly relied upon.



































































![Apple Seeds tvOS 26 Beta 2 to Developers [Download]](https://www.iclarified.com/images/news/97691/97691/97691-640.jpg)
![Apple Seeds watchOS 26 Beta 2 to Developers [Download]](https://www.iclarified.com/images/news/97688/97688/97688-640.jpg)
![Apple Releases iOS 26 Beta 2 and iPadOS 26 Beta 2 [Download]](https://www.iclarified.com/images/news/97685/97685/97685-640.jpg)
![Apple Seeds visionOS 26 Beta 2 to Developers [Download]](https://www.iclarified.com/images/news/97693/97693/97693-640.jpg)









































































































_marcos_alvarado_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)






































































































![[The AI Show Episode 154]: AI Answers: The Future of AI Agents at Work, Building an AI Roadmap, Choosing the Right Tools, & Responsible AI Use](https://www.marketingaiinstitute.com/hubfs/ep%20154%20cover.png)
![[The AI Show Episode 153]: OpenAI Releases o3-Pro, Disney Sues Midjourney, Altman: “Gentle Singularity” Is Here, AI and Jobs & News Sites Getting Crushed by AI Search](https://www.marketingaiinstitute.com/hubfs/ep%20153%20cover.png)


































































































































































.png?#)