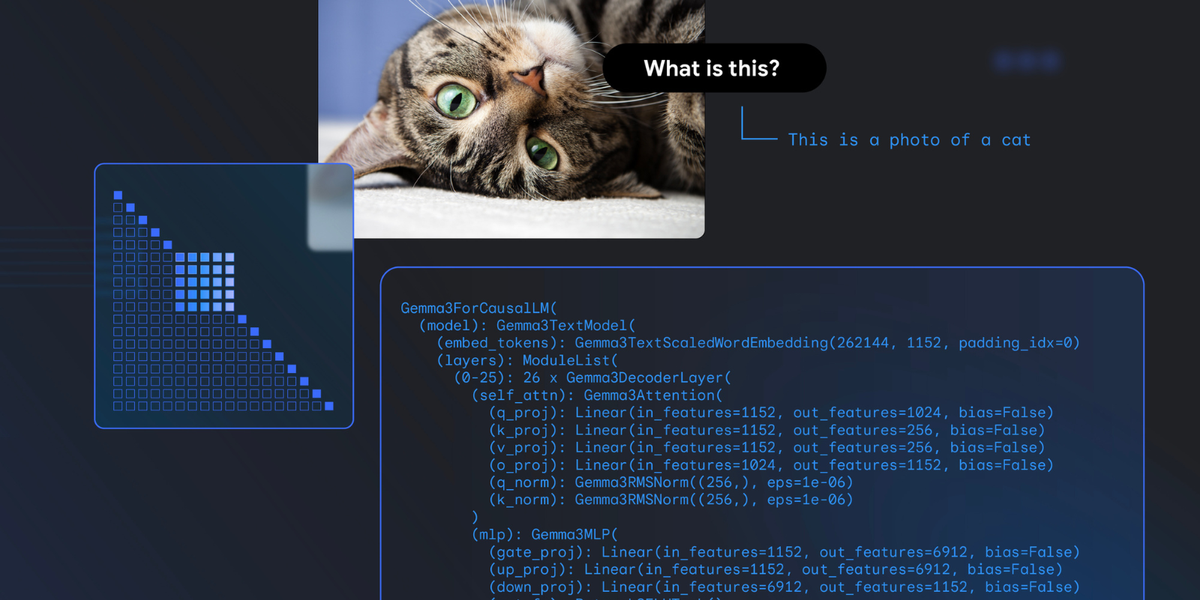
Building a Local RAG System with MCP for VS Code AI Agents: A Technical Deep Dive
In my previous post, I shared how I supercharged my VS Code AI agent with a local RAG (Retrieval-Augmented Generation) system using MCP (Model Context Protocol). Today, I'm following up with a detailed technical tutorial on how I built this system from scratch. This tutorial will walk you through creating a local RAG system that indexes your Markdown journal entries and exposes semantic search capabilities to VS Code AI agents through an MCP server. By the end, you'll have a powerful system that gives your AI assistant "memory" of your past writings. The complete code for this project is available on GitHub: https://github.com/estevaom/md-rag-mcp Prerequisites Python 3.8+ installed NVIDIA GPU with CUDA support (optional but recommended for faster embeddings) VS Code with an AI agent that supports MCP (like Cursor, Cline or Roo Code) Basic understanding of Python and RAG concepts For Windows users, detailed instructions on setting up the environment, including WSL and CUDA, can be found in the install_instructions.md file. Project Structure Before diving into the code, let's understand the project structure: / ├── README.md # Project overview ├── journal/ # Journal entries │ ├── 2025/ # Organized by year │ │ └── 04/ # Month │ │ ├── 18.md # Daily entries │ │ └── 19.md │ └── topics/ # Topic-based entries (future use) ├── code/ # Code and scripts │ ├── mcp/ # MCP server code │ │ └── journal_rag_mcp.py │ ├── scripts/ # Python scripts │ │ └── rag_search.py │ └── data/ # Data storage (e.g., vector DB) │ └── chroma_db/ # Vector database (example) └── .venv/ # Python virtual environment Step 1: Setting Up the Environment First, let's create a virtual environment and install the necessary packages: # Create a virtual environment python -m venv .venv # Activate the virtual environment source .venv/bin/activate # On Windows: .venv\Scripts\activate # Install required packages pip install sentence-transformers chromadb langchain-text-splitters python-frontmatter rich torch If you have an NVIDIA GPU, ensure you have the appropriate CUDA toolkit installed to leverage GPU acceleration for generating embeddings. Step 2: Building the RAG System Let's start by implementing the core RAG functionality in code/scripts/rag_search.py. 2.1 Importing Dependencies import os import frontmatter # For parsing Markdown YAML front matter from langchain_text_splitters import RecursiveCharacterTextSplitter from sentence_transformers import SentenceTransformer import chromadb import torch # For GPU detection from rich.console import Console # For nice printing # Initialize console for rich printing console = Console() 2.2 Configuration # Get the project root directory PROJECT_ROOT = os.path.dirname(os.path.dirname(os.path.dirname(os.path.abspath(__file__)))) # 3 levels up from code/scripts/rag_search.py DATA_DIRECTORY = os.path.join(PROJECT_ROOT, "journal") CHROMA_DB_PATH = os.path.join(PROJECT_ROOT, "data", "chroma_db") CHROMA_COLLECTION_NAME = "life_journal_collection" EMBEDDING_MODEL_NAME = 'all-MiniLM-L6-v2' # Efficient, high-quality embedding model CHUNK_SIZE = 500 # Max characters per chunk CHUNK_OVERLAP = 50 # Characters overlap between chunks 2.3 Loading Documents def load_markdown_docs(directory_path): """ Recursively loads all Markdown files from the specified directory and its subdirectories, parsing YAML front matter. """ documents = [] if not os.path.isdir(directory_path): console.print(f"[bold red]Error: Directory not found:[/bold red] {directory_path}") return documents console.print(f"Scanning directory: [cyan]{directory_path}[/cyan]") found_files = False # Walk through directory and subdirectories for root, dirs, files in os.walk(directory_path): for filename in files: if filename.endswith(".md"): found_files = True filepath = os.path.join(root, filename) # Get relative path from the base directory for better source identification rel_path = os.path.relpath(filepath, directory_path) try: post = frontmatter.load(filepath) # Add filename and path to metadata for later reference if 'source' not in post.metadata: post.metadata['source'] = rel_path documents.append(post) console.print(f" [green]Loaded:[/green] {rel_path}") except Exception as e: console.print(f" [bold red]Error loading {rel_path}:[/bold red] {e}") if not found_files: console.print(f"[yell

In my previous post, I shared how I supercharged my VS Code AI agent with a local RAG (Retrieval-Augmented Generation) system using MCP (Model Context Protocol). Today, I'm following up with a detailed technical tutorial on how I built this system from scratch.
This tutorial will walk you through creating a local RAG system that indexes your Markdown journal entries and exposes semantic search capabilities to VS Code AI agents through an MCP server. By the end, you'll have a powerful system that gives your AI assistant "memory" of your past writings.
The complete code for this project is available on GitHub: https://github.com/estevaom/md-rag-mcp
Prerequisites
- Python 3.8+ installed
- NVIDIA GPU with CUDA support (optional but recommended for faster embeddings)
- VS Code with an AI agent that supports MCP (like Cursor, Cline or Roo Code)
- Basic understanding of Python and RAG concepts
For Windows users, detailed instructions on setting up the environment, including WSL and CUDA, can be found in the
install_instructions.mdfile.
Project Structure
Before diving into the code, let's understand the project structure:
/
├── README.md # Project overview
├── journal/ # Journal entries
│ ├── 2025/ # Organized by year
│ │ └── 04/ # Month
│ │ ├── 18.md # Daily entries
│ │ └── 19.md
│ └── topics/ # Topic-based entries (future use)
├── code/ # Code and scripts
│ ├── mcp/ # MCP server code
│ │ └── journal_rag_mcp.py
│ ├── scripts/ # Python scripts
│ │ └── rag_search.py
│ └── data/ # Data storage (e.g., vector DB)
│ └── chroma_db/ # Vector database (example)
└── .venv/ # Python virtual environment
Step 1: Setting Up the Environment
First, let's create a virtual environment and install the necessary packages:
# Create a virtual environment
python -m venv .venv
# Activate the virtual environment
source .venv/bin/activate # On Windows: .venv\Scripts\activate
# Install required packages
pip install sentence-transformers chromadb langchain-text-splitters python-frontmatter rich torch
If you have an NVIDIA GPU, ensure you have the appropriate CUDA toolkit installed to leverage GPU acceleration for generating embeddings.
Step 2: Building the RAG System
Let's start by implementing the core RAG functionality in code/scripts/rag_search.py.
2.1 Importing Dependencies
import os
import frontmatter # For parsing Markdown YAML front matter
from langchain_text_splitters import RecursiveCharacterTextSplitter
from sentence_transformers import SentenceTransformer
import chromadb
import torch # For GPU detection
from rich.console import Console # For nice printing
# Initialize console for rich printing
console = Console()
2.2 Configuration
# Get the project root directory
PROJECT_ROOT = os.path.dirname(os.path.dirname(os.path.dirname(os.path.abspath(__file__)))) # 3 levels up from code/scripts/rag_search.py
DATA_DIRECTORY = os.path.join(PROJECT_ROOT, "journal")
CHROMA_DB_PATH = os.path.join(PROJECT_ROOT, "data", "chroma_db")
CHROMA_COLLECTION_NAME = "life_journal_collection"
EMBEDDING_MODEL_NAME = 'all-MiniLM-L6-v2' # Efficient, high-quality embedding model
CHUNK_SIZE = 500 # Max characters per chunk
CHUNK_OVERLAP = 50 # Characters overlap between chunks
2.3 Loading Documents
def load_markdown_docs(directory_path):
"""
Recursively loads all Markdown files from the specified directory and its subdirectories,
parsing YAML front matter.
"""
documents = []
if not os.path.isdir(directory_path):
console.print(f"[bold red]Error: Directory not found:[/bold red] {directory_path}")
return documents
console.print(f"Scanning directory: [cyan]{directory_path}[/cyan]")
found_files = False
# Walk through directory and subdirectories
for root, dirs, files in os.walk(directory_path):
for filename in files:
if filename.endswith(".md"):
found_files = True
filepath = os.path.join(root, filename)
# Get relative path from the base directory for better source identification
rel_path = os.path.relpath(filepath, directory_path)
try:
post = frontmatter.load(filepath)
# Add filename and path to metadata for later reference
if 'source' not in post.metadata:
post.metadata['source'] = rel_path
documents.append(post)
console.print(f" [green]Loaded:[/green] {rel_path}")
except Exception as e:
console.print(f" [bold red]Error loading {rel_path}:[/bold red] {e}")
if not found_files:
console.print(f"[yellow]Warning: No .md files found in {directory_path} or its subdirectories[/yellow]")
return documents
2.4 Splitting Documents into Chunks
def split_docs(documents):
"""
Splits the content of loaded documents into smaller chunks.
"""
console.print("\nSplitting documents into chunks...")
text_splitter = RecursiveCharacterTextSplitter(
chunk_size=CHUNK_SIZE,
chunk_overlap=CHUNK_OVERLAP,
length_function=len,
is_separator_regex=False,
)
all_chunks = []
for doc in documents:
chunks = text_splitter.split_text(doc.content)
for i, chunk_text in enumerate(chunks):
# Create a unique ID for the chunk based on source and index
chunk_id = f"{doc.metadata.get('source', 'unknown')}_{i}"
chunk_metadata = doc.metadata.copy() # Start with original metadata
chunk_metadata['chunk_index'] = i
chunk_metadata['chunk_id'] = chunk_id # Store ID in metadata too
all_chunks.append({
"id": chunk_id, # ID for ChromaDB
"text": chunk_text,
"metadata": chunk_metadata
})
console.print(f" Split into {len(all_chunks)} chunks.")
return all_chunks
2.5 Initializing the Embedding Model
def initialize_embedding_model(model_name):
"""Initializes and returns the Sentence Transformer model with GPU support if available."""
console.print(f"\nInitializing embedding model: [cyan]{model_name}[/cyan]")
try:
# Check if CUDA is available
device = "cuda" if torch.cuda.is_available() else "cpu"
if device == "cuda":
console.print(f" [green]CUDA is available! Using GPU: {torch.cuda.get_device_name(0)}[/green]")
else:
console.print(" [yellow]CUDA not available. Using CPU for embeddings (slower).[/yellow]")
# Load model with device specification
model = SentenceTransformer(model_name, device=device)
console.print(" [green]Embedding model loaded successfully.[/green]")
return model
except Exception as e:
console.print(f"[bold red]Error initializing embedding model:[/bold red] {e}")
return None
2.6 Setting Up the Vector Database
def initialize_vector_store(db_path, collection_name):
"""Initializes and returns the ChromaDB client and collection."""
console.print(f"\nInitializing vector store at: [cyan]{db_path}[/cyan]")
try:
chroma_client = chromadb.PersistentClient(path=db_path)
# Get the collection if it exists, or create it if it doesn't
collection = chroma_client.get_or_create_collection(name=collection_name)
console.print(f" [green]Ensured vector store collection '{collection_name}' exists.[/green]")
return collection
except Exception as e:
console.print(f"[bold red]Error initializing vector store:[/bold red] {e}")
return None
2.7 Indexing Chunks
def index_chunks(collection, chunks, embedding_model):
"""Generates embeddings and indexes chunks in the vector store."""
if not collection or not chunks or not embedding_model:
console.print("[bold red]Error: Cannot index chunks due to missing components.[/bold red]")
return False
console.print(f"\nIndexing {len(chunks)} chunks...")
# Prepare data for ChromaDB batch insertion
ids = [chunk['id'] for chunk in chunks]
documents = [chunk['text'] for chunk in chunks]
# Process metadata to ensure all values are of compatible types (str, int, float, bool)
processed_metadatas = []
for chunk in chunks:
processed_metadata = {}
for key, value in chunk['metadata'].items():
# Convert any non-compatible types to strings
if isinstance(value, (str, int, float, bool)):
processed_metadata[key] = value
else:
processed_metadata[key] = str(value)
processed_metadatas.append(processed_metadata)
metadatas = processed_metadatas
try:
console.print(" Generating embeddings (this may take a moment)...")
embeddings = embedding_model.encode(documents, show_progress_bar=True)
console.print(" Embeddings generated.")
console.print(f" Adding {len(ids)} items to collection '{collection.name}'...")
# Use upsert to add new or update existing chunks by ID
collection.upsert(
ids=ids,
embeddings=embeddings.tolist(), # Convert numpy array to list
documents=documents,
metadatas=metadatas
)
console.print(" [green]Chunks indexed successfully.[/green]")
return True
except Exception as e:
console.print(f"[bold red]Error during indexing:[/bold red] {e}")
return False
2.8 Querying the Vector Database
def query_rag(collection, embedding_model, query_text, n_results=3):
"""
Queries the vector store for chunks similar to the query text.
"""
if not collection or not embedding_model or not query_text:
console.print("[bold red]Error: Cannot query due to missing components.[/bold red]")
return None
try:
console.print(f"\n Generating embedding for query: '{query_text}'")
# Ensure query_text is handled correctly if empty or invalid
if not isinstance(query_text, str) or not query_text.strip():
console.print("[yellow]Warning: Empty query provided.[/yellow]")
return None
query_embedding = embedding_model.encode([query_text.strip()]) # Encode expects a list
console.print(f" Querying collection '{collection.name}' for {n_results} results...")
results = collection.query(
query_embeddings=query_embedding.tolist(), # Convert numpy array to list
n_results=n_results,
include=['documents', 'metadatas', 'distances'] # Include distances for relevance check
)
console.print(" [green]Query successful.[/green]")
return results
except Exception as e:
# Log the full exception for debugging
import traceback
console.print(f"[bold red]Error during query:[/bold red]\n{traceback.format_exc()}")
return None
2.9 Main Execution
if __name__ == "__main__":
console.print("[bold blue]=== Starting Journal RAG Search ===[/bold blue]")
# 1. Load Documents
loaded_docs = load_markdown_docs(DATA_DIRECTORY)
if not loaded_docs:
console.print("[bold red]No documents loaded. Exiting.[/bold red]")
exit() # Exit if loading failed
console.print(f"\n[bold green]Successfully loaded {len(loaded_docs)} documents.[/bold green]")
# 2. Split Documents
doc_chunks = split_docs(loaded_docs)
if not doc_chunks:
console.print("[bold red]Failed to split documents into chunks. Exiting.[/bold red]")
exit() # Exit if splitting failed
# 3. Initialize Models & Vector Store
embed_model = initialize_embedding_model(EMBEDDING_MODEL_NAME)
chroma_collection = initialize_vector_store(CHROMA_DB_PATH, CHROMA_COLLECTION_NAME)
if not embed_model or not chroma_collection:
console.print("[bold red]Failed to initialize models or vector store. Exiting.[/bold red]")
exit() # Exit if initialization failed
# 4. Index Chunks
indexing_successful = index_chunks(chroma_collection, doc_chunks, embed_model)
if indexing_successful:
console.print("\n[bold green]Indexing complete.[/bold green]")
# 5. Interactive Querying
console.print("\n--- Query Test ---")
while True:
query = input("Enter your query (or type 'quit' to exit): ")
if query.lower() == 'quit':
break
if not query:
continue
results = query_rag(chroma_collection, embed_model, query, n_results=3)
console.print("\n[bold magenta]Query Results:[/bold magenta]")
if results and results.get('ids') and results['ids'] and results['ids'][0]:
if not results.get('documents') or not results['documents'][0]:
console.print(" No relevant results found.")
continue
for i, res_doc in enumerate(results['documents'][0]):
distance = results.get('distances', [[None]])[0][i]
metadata = results.get('metadatas', [[{}]])[0][i]
source = metadata.get('source', 'N/A')
distance_str = f"{distance:.4f}" if distance is not None else "N/A"
console.print(f" [cyan]Result {i+1} (Distance: {distance_str}):[/cyan]")
console.print(f" [dim]Source: {source}[/dim]")
console.print(f" {res_doc}")
elif results is None:
console.print(" An error occurred during the query.")
else:
console.print(" No relevant results found.")
else:
console.print("[bold red]Indexing failed. Check errors above.[/bold red]")
console.print("\n[bold blue]=== Journal RAG Search Finished ===[/bold blue]")
Step 3: Creating the MCP Server
Now, let's implement the MCP server that will expose our RAG system to VS Code AI agents. Create a file at code/mcp/journal_rag_mcp.py:
3.1 Imports and Configuration
#!/usr/bin/env python3
import sys
import json
import os
import time
import traceback
import frontmatter
# --- Early Startup Logging ---
print("[MCP Server Debug] Script starting up", file=sys.stderr, flush=True)
print(f"[MCP Server Debug] Python version: {sys.version}", file=sys.stderr, flush=True)
print(f"[MCP Server Debug] Current working directory: {os.getcwd()}", file=sys.stderr, flush=True)
print(f"[MCP Server Debug] Script directory: {os.path.dirname(os.path.abspath(__file__))}", file=sys.stderr, flush=True)
try:
import chromadb
from sentence_transformers import SentenceTransformer
import torch
except ImportError as e:
print(json.dumps({
"jsonrpc": "2.0",
"error": {
"code": -32000, # Server error
"message": f"Missing required Python package: {e}. Please install chromadb, sentence-transformers, and torch.",
}
}), file=sys.stderr)
sys.exit(1)
# --- Configuration ---
# Calculate PROJECT_ROOT based on this script's location (code/mcp/journal_rag_mcp.py)
SCRIPT_DIR = os.path.dirname(os.path.abspath(__file__))
PROJECT_ROOT = os.path.dirname(os.path.dirname(SCRIPT_DIR)) # 2 levels up
CHROMA_DB_PATH = os.path.join(PROJECT_ROOT, "data", "chroma_db")
CHROMA_COLLECTION_NAME = "life_journal_collection"
CHROMA_COLLECTION_NAME = "life_journal_collection"
EMBEDDING_MODEL_NAME = 'all-MiniLM-L6-v2'
DEFAULT_N_RESULTS = 3
# --- Global Variables ---
embedding_model = None
chroma_collection = None
is_initialized = False
3.2 Logging and Initialization
# --- Logging ---
def log_error(message):
"""Logs an error message to stderr."""
print(f"[MCP Server Error] {message}", file=sys.stderr, flush=True)
def log_info(message):
"""Logs an info message to stderr."""
print(f"[MCP Server Info] {message}", file=sys.stderr, flush=True)
# --- Initialization Function ---
def initialize_resources():
"""Loads the embedding model and connects to ChromaDB."""
global embedding_model, chroma_collection, is_initialized
if is_initialized:
return True
log_info("Initializing resources...")
try:
# 1. Initialize Embedding Model
log_info(f"Loading embedding model: {EMBEDDING_MODEL_NAME}")
device = "cuda" if torch.cuda.is_available() else "cpu"
log_info(f"Using device: {device}")
embedding_model = SentenceTransformer(EMBEDDING_MODEL_NAME, device=device)
log_info("Embedding model loaded.")
# 2. Initialize ChromaDB
log_info(f"Connecting to ChromaDB at: {CHROMA_DB_PATH}")
if not os.path.exists(CHROMA_DB_PATH):
log_info(f"ChromaDB path does not exist: {CHROMA_DB_PATH}. Creating it now.")
try:
os.makedirs(CHROMA_DB_PATH, exist_ok=True)
log_info(f"Created ChromaDB directory: {CHROMA_DB_PATH}")
except Exception as e:
log_error(f"Failed to create ChromaDB directory: {e}")
log_error(traceback.format_exc())
return False # Indicate initialization failure
chroma_client = chromadb.PersistentClient(path=CHROMA_DB_PATH)
log_info(f"Getting/Creating collection: {CHROMA_COLLECTION_NAME}")
chroma_collection = chroma_client.get_or_create_collection(name=CHROMA_COLLECTION_NAME)
log_info("ChromaDB collection ready.")
is_initialized = True
log_info("Resources initialized successfully.")
return True
except Exception as e:
log_error(f"Initialization failed: {e}")
log_error(traceback.format_exc())
embedding_model = None
chroma_collection = None
is_initialized = False
return False
3.3 Timestamp Tracking for Incremental Updates
# --- Timestamp Tracking Functions ---
def get_last_indexed_time():
"""Gets the timestamp of the last indexing operation."""
timestamp_file = os.path.join(PROJECT_ROOT, "data", "last_indexed_time.txt")
if not os.path.exists(timestamp_file):
return 0 # Return 0 if file doesn't exist (never indexed)
try:
with open(timestamp_file, 'r') as f:
timestamp = float(f.read().strip())
return timestamp
except:
return 0 # Return 0 if there's an error reading the file
def update_last_indexed_time():
"""Updates the timestamp of the last indexing operation to current time."""
timestamp_file = os.path.join(PROJECT_ROOT, "data", "last_indexed_time.txt")
try:
# Create directory if it doesn't exist
timestamp_dir = os.path.dirname(timestamp_file)
log_info(f"Ensuring directory exists: {timestamp_dir}")
os.makedirs(timestamp_dir, exist_ok=True)
# Write current timestamp
current_time = time.time()
log_info(f"Writing timestamp {current_time} to {timestamp_file}")
with open(timestamp_file, 'w') as f:
f.write(str(current_time))
log_info(f"Timestamp updated successfully")
return True
except Exception as e:
log_error(f"Error updating timestamp: {e}")
log_error(traceback.format_exc())
return False
def load_markdown_docs_since(directory_path, timestamp):
"""
Loads only markdown files that have been modified since the given timestamp.
"""
documents = []
if not os.path.isdir(directory_path):
log_error(f"Error: Directory not found: {directory_path}")
return documents
log_info(f"Scanning directory for modified files: {directory_path}")
found_files = False
# Walk through directory and subdirectories
for root, dirs, files in os.walk(directory_path):
for filename in files:
if filename.endswith(".md"):
filepath = os.path.join(root, filename)
# Check if file was modified after the timestamp
if os.path.getmtime(filepath) > timestamp:
found_files = True
# Get relative path from the base directory for better source identification
rel_path = os.path.relpath(filepath, directory_path)
try:
post = frontmatter.load(filepath)
# Add filename and path to metadata for later reference
if 'source' not in post.metadata:
post.metadata['source'] = rel_path
documents.append(post)
log_info(f" Loaded modified file: {rel_path}")
except Exception as e:
log_error(f" Error loading {rel_path}: {e}")
if not found_files:
log_info(f"No modified .md files found since {time.ctime(timestamp)}")
return documents
3.4 Index Update Function
# --- Index Update Function ---
def update_journal_index(full_reindex=False):
"""
Updates the journal index with new or modified entries.
"""
try:
# Import functions from rag_search.py
sys.path.append(os.path.join(PROJECT_ROOT, "code", "scripts"))
try:
from rag_search import (
load_markdown_docs, split_docs, initialize_embedding_model,
initialize_vector_store, index_chunks
)
except ImportError as e:
log_error(f"Error importing from rag_search.py: {e}")
log_error(f"sys.path: {sys.path}")
return False, f"Error importing from rag_search.py: {e}"
# Define data directory (same as in rag_search.py)
DATA_DIRECTORY = os.path.join(PROJECT_ROOT, "journal")
# Track stats for reporting
files_processed = 0
chunks_indexed = 0
# Load documents (all or only new/modified)
if full_reindex:
# Process all documents
log_info("Performing full reindex of all journal entries")
documents = load_markdown_docs(DATA_DIRECTORY)
files_processed = len(documents)
else:
# Get last indexing timestamp
last_indexed_time = get_last_indexed_time()
log_info(f"Performing incremental index update since {time.ctime(last_indexed_time)}")
# Process only new or modified documents
documents = load_markdown_docs_since(DATA_DIRECTORY, last_indexed_time)
files_processed = len(documents)
if not documents:
return True, "No new or modified journal entries found."
log_info(f"Found {files_processed} files to process")
# Split documents into chunks
chunks = split_docs(documents)
chunks_indexed = len(chunks)
if not chunks:
return True, f"No chunks generated from {files_processed} files."
log_info(f"Split into {chunks_indexed} chunks")
# Initialize embedding model and vector store
embed_model = initialize_embedding_model(EMBEDDING_MODEL_NAME)
chroma_collection = initialize_vector_store(CHROMA_DB_PATH, CHROMA_COLLECTION_NAME)
if not embed_model or not chroma_collection:
return False, "Failed to initialize embedding model or vector store."
# Index chunks
log_info(f"Indexing {chunks_indexed} chunks")
indexing_successful = index_chunks(chroma_collection, chunks, embed_model)
if indexing_successful:
# Update last indexed time
update_last_indexed_time()
return True, f"Successfully indexed {chunks_indexed} chunks from {files_processed} files."
else:
return False, "Indexing failed."
except Exception as e:
log_error(f"Error updating index: {str(e)}")
log_error(traceback.format_exc())
return False, f"Error updating index: {str(e)}"
3.5 RAG Query Function
# --- RAG Query Function ---
def perform_rag_query(query_text: str, n_results: int = DEFAULT_N_RESULTS):
"""Executes a semantic search query."""
if not is_initialized or embedding_model is None or chroma_collection is None:
log_error("Query attempted before resources were initialized.")
return None, "Resources not initialized"
try:
log_info(f"Encoding query: '{query_text}'")
query_embedding = embedding_model.encode([query_text.strip()])
log_info(f"Querying collection '{chroma_collection.name}' for {n_results} results...")
results = chroma_collection.query(
query_embeddings=query_embedding.tolist(),
n_results=n_results,
include=['documents', 'metadatas', 'distances']
)
log_info("Query successful.")
# Format results
formatted_results = []
if results and results.get('ids') and results['ids'][0]:
for i, doc_text in enumerate(results['documents'][0]):
metadata = results['metadatas'][0][i] if results['metadatas'] and results['metadatas'][0] else {}
distance = results['distances'][0][i] if results['distances'] and results['distances'][0] else None
formatted_results.append({
"source": metadata.get('source', 'N/A'),
"text": doc_text,
"distance": distance
})
return formatted_results, None # Results, No error message
except Exception as e:
log_error(f"Error during query: {e}")
log_error(traceback.format_exc())
return None, str(e) # No results, Error message
3.6 MCP Response Formatting
# --- MCP Response Formatting ---
def create_mcp_response(request_id, result=None, error=None):
"""Creates a JSON-RPC 2.0 response dictionary."""
response = {"jsonrpc": "2.0", "id": request_id}
if error:
response["error"] = error
else:
response["result"] = result
return response
def create_mcp_error(code, message):
"""Creates an MCP error object."""
return {"code": code, "message": message}
3.7 Main Server Loop
# --- Main Server Loop ---
def main():
"""Reads MCP requests from stdin, processes them, writes responses to stdout."""
if not initialize_resources():
# Send an error response if init fails
log_error("Exiting due to initialization failure.")
sys.exit(1) # Exit if resources can't be loaded
log_info("MCP server started. Waiting for requests on stdin...")
for line in sys.stdin:
request_id = None # Reset for each request
try:
log_info(f"Received request line: {line.strip()}")
request = json.loads(line)
request_id = request.get("id")
method = request.get("method")
params = request.get("params", {})
log_info(f"Processing method: {method} for request ID: {request_id}")
response = None
if method == "initialize":
log_info("Handling 'initialize' method.")
# Handle the initial handshake from the AI agent
response = create_mcp_response(request_id, result={
"protocolVersion": "2024-11-05", # Specify MCP protocol version
"serverInfo": {
"name": "journal-rag-mcp-server", # Required server name
"displayName": "Journal RAG Server",
"version": "0.1.0",
},
"capabilities": {
"tools": {
"listChanged": False # We don't support notifications for tool list changes
}
}
})
elif method == "notifications/initialized":
# This is just a notification, no response needed
log_info("Received initialized notification")
response = None # No response needed for notifications
elif method == "tools/list":
log_info("Handling 'tools/list' method.")
response = create_mcp_response(request_id, result={
"tools": [
{
"name": "query_journal",
"description": "Queries the personal journal entries using semantic search.",
"inputSchema": {
"type": "object",
"properties": {
"query": {
"type": "string",
"description": "The search query text."
},
"n_results": {
"type": "number",
"description": f"Number of results to return (default: {DEFAULT_N_RESULTS}).",
"minimum": 1
}
},
"required": ["query"]
}
},
{
"name": "update_index",
"description": "Updates the journal index with new or modified entries",
"inputSchema": {
"type": "object",
"properties": {
"full_reindex": {
"type": "boolean",
"description": "Whether to reindex all journal entries (true) or only new/modified ones (false)",
"default": false
}
}
}
}
]
})
elif method == "resources/list":
log_info("Handling 'resources/list' method.")
# Return empty list if we don't have resources
response = create_mcp_response(request_id, result={
"resources": []
})
elif method == "resources/templates/list":
log_info("Handling 'resources/templates/list' method.")
# Return empty list if we don't have resource templates
response = create_mcp_response(request_id, result={
"templates": []
})
elif method == "tools/call":
log_info("Handling 'tools/call' method.")
tool_name = params.get("name")
arguments = params.get("arguments", {})
log_info(f"Tool name: {tool_name}, Arguments: {arguments}")
if tool_name == "query_journal":
query = arguments.get("query")
n_results = arguments.get("n_results", DEFAULT_N_RESULTS)
if not query or not isinstance(query, str):
response = create_mcp_response(request_id, error=create_mcp_error(-32602, "Invalid params: 'query' argument is missing or not a string."))
elif not isinstance(n_results, int) or n_results < 1:
response = create_mcp_response(request_id, error=create_mcp_error(-32602, f"Invalid params: 'n_results' must be a positive integer (default: {DEFAULT_N_RESULTS})."))
else:
results, error_msg = perform_rag_query(query, n_results)
if error_msg:
response = create_mcp_response(request_id, error=create_mcp_error(-32000, f"Query execution failed: {error_msg}"))
else:
# Embed the list of results as a JSON string within the text content
response = create_mcp_response(request_id, result={
"content": [{
"type": "text",
"text": json.dumps(results, indent=2) # Pretty print for readability if needed
}]
})
elif tool_name == "update_index":
full_reindex = arguments.get("full_reindex", False)
log_info(f"Updating index with full_reindex={full_reindex}")
success, message = update_journal_index(full_reindex)
if success:
response = create_mcp_response(request_id, result={
"content": [{
"type": "text",
"text": message
}]
})
else:
response = create_mcp_response(
request_id,
error=create_mcp_error(-32000, f"Index update failed: {message}")
)
else:
log_error(f"Unknown tool called: {tool_name}")
response = create_mcp_response(request_id, error=create_mcp_error(-32601, f"Method not found: Unknown tool '{tool_name}'"))
else:
log_error(f"Unknown method received: {method}")
response = create_mcp_response(request_id, error=create_mcp_error(-32601, f"Method not found: Unknown method '{method}'"))
if response:
response_json = json.dumps(response)
log_info(f"Prepared response for ID {request_id}: {response_json[:200]}...") # Log truncated response
print(response_json, flush=True)
log_info(f"Successfully sent response for request ID: {request_id}")
elif method == "notifications/initialized":
# This is expected for notifications that don't require responses
log_info(f"No response needed for notification: {method}")
else:
log_error(f"No response generated for request ID: {request_id}, method: {method}")
except json.JSONDecodeError:
log_error(f"JSONDecodeError: Failed to decode JSON request: {line.strip()}")
error_response = create_mcp_response(request_id, error=create_mcp_error(-32700, "Parse error: Invalid JSON received."))
print(json.dumps(error_response), flush=True)
except Exception as e:
log_error(f"Unexpected error processing request: {e}")
log_error(traceback.format_exc())
error_response = create_mcp_response(request_id, error=create_mcp_error(-32000, f"Internal server error: {e}"))
print(json.dumps(error_response), flush=True)
# --- Script Execution Guard ---
if __name__ == "__main__":
try:
main()
except KeyboardInterrupt:
log_info("Server stopped by KeyboardInterrupt.")
except Exception as e:
log_error(f"Unhandled exception in main execution: {e}")
log_error(traceback.format_exc())
sys.exit(1) # Ensure non-zero exit code on unhandled error
finally:
log_info("Server process ending.")
Step 4: Integrating with VS Code
Now that we have our RAG system and MCP server implemented, we need to integrate it with VS Code. This involves configuring VS Code to recognize and connect to our MCP server.
4.1 Configure the MCP Server in VS Code Settings
To connect VS Code to your Journal RAG MCP server, you need to add its configuration to your VS Code settings. Open your VS Code settings (File > Preferences > Settings) and search for "MCP Servers". Add the following JSON configuration:
{
"mcpServers": {
"journal-rag-mcp": {
"command": "${workspaceFolder}/.venv/bin/python3",
"args": [
"./code/mcp/journal_rag_mcp.py" # Path relative to workspaceFolder
],
"env": {},
"disabled": false,
"alwaysAllow": [],
"autoApprove": [
"query_journal"
]
}
}
}
This configuration tells VS Code how to start your MCP server. The ${workspaceFolder} variable is a built-in VS Code variable that resolves to the path of your currently open workspace folder (which should be the project root).
After adding this configuration, restart VS Code for the changes to take effect. Your AI agent should now be able to detect and interact with the "journal-rag-mcp" server.
This configuration tells VS Code's AI assistant (like Cline or Roo Code) to connect to our MCP server when it starts up.
Step 5: Testing and Usage
Now that everything is set up, let's test our RAG system and MCP server.
5.1 Running the RAG System Standalone
You can run the RAG system standalone to test indexing and querying:
# Activate the virtual environment
source .venv/bin/activate
# Run the RAG search script
python code/scripts/rag_search.py
This will index all your journal entries and allow you to interactively query them.
5.2 Using the MCP Server with VS Code
- Open VS Code in your project directory
- Start a conversation with your AI assistant (Cline or Roo Code)
- The MCP server will automatically start and connect
- You can now ask your AI assistant questions about your journal entries
Example queries:
- "What did I write about anxiety last month?"
- "Summarize my thoughts on my work project from last week"
- "Find entries where I discussed my relationship with my mother"
The AI assistant will use the query_journal tool to retrieve relevant information from your journal entries and provide a response based on that information.
Step 6: Advanced Features
6.1 Incremental Indexing
Our implementation includes incremental indexing, which only processes files that have been modified since the last indexing operation. This makes the indexing process much faster when you only add or modify a few files.
To trigger an incremental update:
Update the rag-mcp index.
Okay, I will update the journal index now.
Cline wants to use a tool on the journal-rag-mcp MCP server:
update_index
Updates the journal index with new or modified entries
Arguments
{
"full_reindex": false
}
6.2 GPU Acceleration
If you have an NVIDIA GPU, our implementation automatically uses it for generating embeddings, which significantly speeds up the indexing and querying process. The code detects if CUDA is available and uses it if possible.
Conclusion
In this tutorial, we've built a powerful local RAG system that integrates with VS Code AI agents through the Model Context Protocol. This system allows your AI assistant to have "memory" of your past journal entries, making it much more effective as a reflective journaling partner.
The key components we've implemented are:
- A RAG system that indexes Markdown journal entries and provides semantic search capabilities
- An MCP server that exposes the RAG system to VS Code AI agents
- Integration with VS Code to connect the AI agent to our MCP server
This implementation is just the beginning. You can extend it in many ways, such as:
- Adding more sophisticated chunking strategies
- Implementing metadata filtering (e.g., by date, tags, or mood)
- Creating a dashboard to visualize journal metrics
- Adding support for other file formats
I hope this tutorial helps you build your own local RAG system and supercharge your VS Code AI agent!


































































![Apple's New Ads Spotlight Apple Watch for Kids [Video]](https://www.iclarified.com/images/news/97197/97197/97197-640.jpg)














![Here’s the Pebble smartwatch reboot in action, and how much tariffs might cost you [Video]](https://i0.wp.com/9to5google.com/wp-content/uploads/sites/4/2025/03/core-2-duo-smartwatch-3.jpg?resize=1200%2C628&quality=82&strip=all&ssl=1)
































































































_Andy_Dean_Photography_Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)



















































































































![[The AI Show Episode 145]: OpenAI Releases o3 and o4-mini, AI Is Causing “Quiet Layoffs,” Executive Order on Youth AI Education & GPT-4o’s Controversial Update](https://www.marketingaiinstitute.com/hubfs/ep%20145%20cover.png)