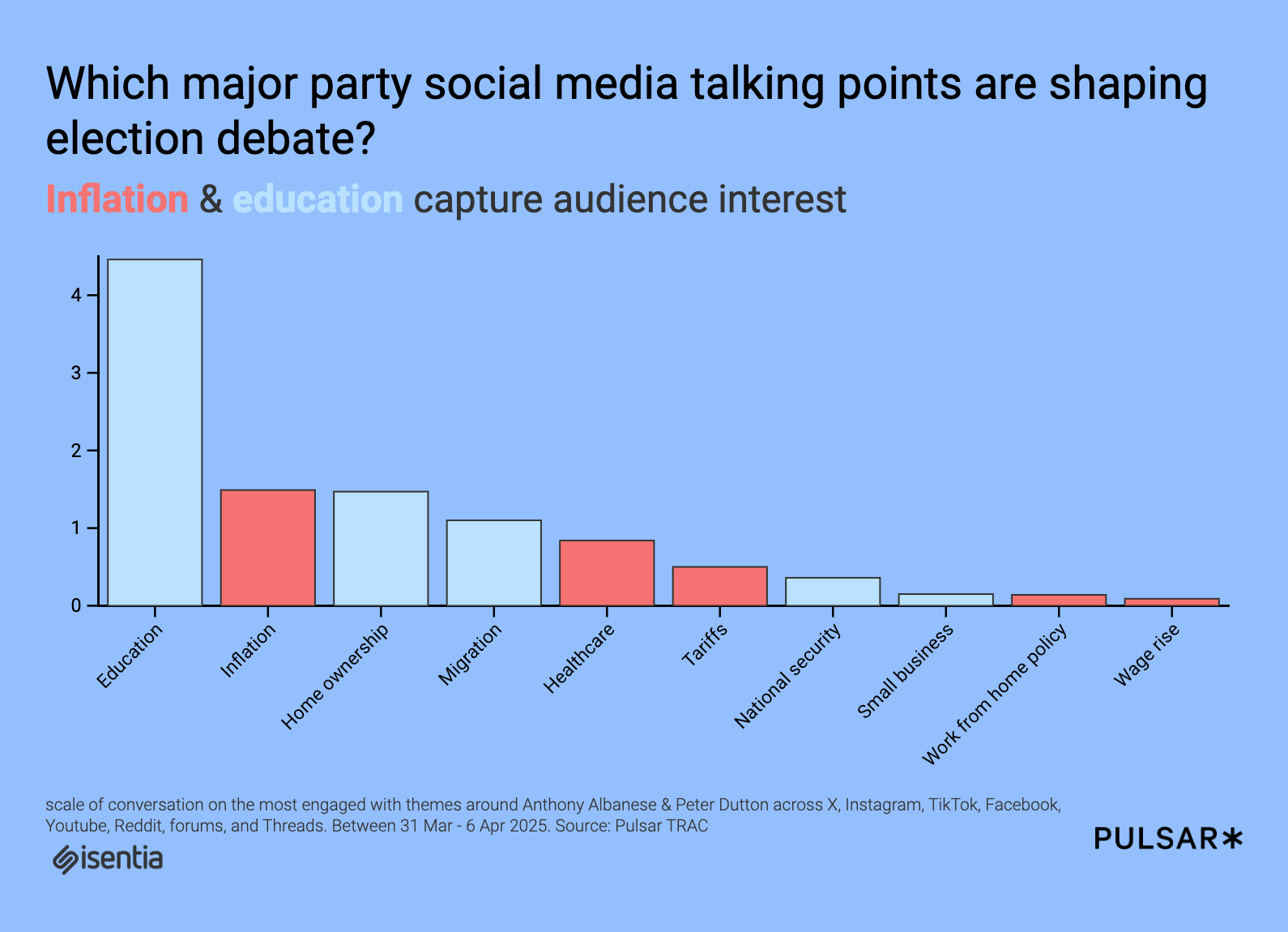
Want Better Clusters? Try DeepType
A smarter way to cluster data using deep learning The post Want Better Clusters? Try DeepType appeared first on Towards Data Science.

As it turns out, Deep Learning can be incredibly useful for clustering problems. Here’s the key idea: suppose we train a neural network using a loss function that reflects something we care about — say, how well we can classify or separate examples. If the network achieves low loss, we can infer that the representations it learns (especially in the second-to-last layer) capture meaningful structure in the data. In other words, these intermediate representations encode what the network has learned about the task.
So, what happens if we run a clustering algorithm (like KMeans) on those representations? Ideally, we end up with clusters that reflect the same underlying structure the network was trained to capture.
Ahh, that’s a lot! Here’s a picture:

As seen in the image, when we run our inputs through until the second-to-last layer, we get a vector out with Kₘ values, which is presumably a lot lower than the amount of inputs we started with if we did everything right. Because the output layer solely looks at this vector when making predictions, if our predictions are good, we can conclude that this vector encapsulates some important information about our data. Clustering in this space is more meaningful than clustering raw data, since we’ve filtered for the features that actually matter.
This is the fundamental idea behind DeepType — a Neural Network approach to clustering. Rather than clustering raw data directly, DeepType first learns a task-relevant representation through supervised training and then performs clustering in that learned space.
This does raise a question, however — if we already have ground-truth labels, why would we need to run clustering? After all, if we just clustered using our labels, wouldn’t that create a perfect clustering? Then, for new data points, we could simply run our neural net, predict the label, and cluster the point appropriately.
As it turns out, in some contexts, we care more about the relationships between our data points than the labels themselves. In the paper that introduced DeepType, for instance, the authors used the idea described to find different groupings of patients with breast cancer based on genetic data, which is very useful in a biological context. They then found that these groups correlated very highly to survival rates, which makes sense given that the representations they clustered on were ingrained with biological knowledge¹.
Refining the Idea: DeepType’s Loss Function
At this point, we understand the core idea: train a neural network to learn a task-relevant representation, then cluster in that space. However, we can make some slight modifications to make this process better.
For starters, we’d like the clusters that we produce to be compact if possible. In other words, we’d much rather have the situation in the picture on the left than on the right:

In order to do this, we want to push the representations of data points in the same clusters to be as close together as possible. To do this, we add a term to our loss function that penalizes the distance between our input’s representation and the center of the cluster its been assigned to. Thus, our loss function becomes

Where d is a distance function between vectors, i.e. the square of the norm of the difference between the vectors (as is used in the original paper).
But wait, how do we get the cluster centers if we haven’t trained the network yet? In order to get around that, DeepType does the following procedure:
- Train a model on just the primary loss
- Create clusters in the representation space (using e.g. KMeans or your favorite algorithm)
- Train the model using the modified loss
- Go back to step 2 and repeat until we converge
Eventually, this procedure produces compact clusters that hopefully correspond to our loss of interest.
Finding Important Inputs
In contexts where DeepType is useful, in addition to caring about clusters, we also care about which inputs are the most informative/important. The paper that introduced DeepType, for instance, was interested in determining which genes were the most important in determining someone’s cancer subtype — such information is certainly useful for a biologist. Plenty of other contexts would also find such information interesting — in fact, it’s hard to dream up one that wouldn’t.
In a deep learning context, we can consider an input to be important if the magnitude of the weights assigned to it by the nodes in the first layer are high. In contrast, if most of our nodes have a weight close to 0 for the input, it won’t contribute much to our final prediction, and hence likely isn’t all that important.
We thus introduce one final loss term — a sparsity loss — that will encourage our neural net to push as many input weights to 0 as possible. With that, our final modified DeepType loss becomes

Where the beta term is the distance term we had before, and the alpha term effectively penalizes a high “magnitude” of the first-layer weight matrix².
We also modify the four-step procedure from the previous section slightly. Instead of just training on the MSE in the first step, we train on both the MSE and the sparsity loss in the pretraining step. Per the authors, our final DeepType structure looks like this:

Playing with DeepType
As part of my research, I’ve posted an open-source implementation of DeepType here. You can additionally download it from pip by doing pip install torch-deeptype .
The DeepType package uses a fairly simple infrastructure to get everything tested. As an example, we’ll create a synthetic dataset with four clusters and 20 inputs, only 5 of which actually contribute to the output:
import numpy as np
import torch
from torch.utils.data import TensorDataset, DataLoader
# 1) Configuration
n_samples = 1000
n_features = 20
n_informative = 5 # number of "important" features
n_clusters = 4 # number of ground-truth clusters
noise_features = n_features - n_informative
# 2) Create distinct cluster centers in the informative subspace
# (spread out so clusters are well separated)
informative_centers = np.random.randn(n_clusters, n_informative) * 5
# 3) Assign each sample to a cluster, then sample around that center
X_informative = np.zeros((n_samples, n_informative))
y_clusters = np.random.randint(0, n_clusters, size=n_samples)
for i, c in enumerate(y_clusters):
center = informative_centers[c]
X_informative[i] = center + np.random.randn(n_informative)
# 4) Generate pure noise for the remaining features
X_noise = np.random.randn(n_samples, noise_features)
# 5) Concatenate informative + noise features
X = np.hstack([X_informative, X_noise]) # shape (1000, 20)
y = y_clusters # shape (1000,)
# 6) Convert to torch tensors and build DataLoader
X_tensor = torch.from_numpy(X).float()
y_tensor = torch.from_numpy(y).long()
dataset = TensorDataset(X_tensor, y_tensor)
train_loader = DataLoader(dataset, batch_size=64, shuffle=True)Here’s what our data looks like when we plot a PCA:

We’ll then define a DeeptypeModel — It can be any infrastructure as long as it implements the forward , get_input_layer_weights , and get_hidden_representations functions:
import torch
import torch.nn as nn
from torch_deeptype import DeeptypeModel
class MyNet(DeeptypeModel):
def __init__(self, input_dim: int, hidden_dim: int, output_dim: int):
super().__init__()
self.input_layer = nn.Linear(input_dim, hidden_dim)
self.h1 = nn.Linear(hidden_dim, hidden_dim)
self.cluster_layer = nn.Linear(hidden_dim, hidden_dim // 2)
self.output_layer = nn.Linear(hidden_dim // 2, output_dim)
def forward(self, x: torch.Tensor) -> torch.Tensor:
# Notice how forward() gets the hidden representations
hidden = self.get_hidden_representations(x)
return self.output_layer(hidden)
def get_input_layer_weights(self) -> torch.Tensor:
return self.input_layer.weight
def get_hidden_representations(self, x: torch.Tensor) -> torch.Tensor:
x = torch.relu(self.input_layer(x))
x = torch.relu(self.h1(x))
x = torch.relu(self.cluster_layer(x))
return xThen, we create a DeeptypeTrainer and train:
from torch_deeptype import DeeptypeTrainer
trainer = DeeptypeTrainer(
model = MyNet(input_dim=20, hidden_dim=64, output_dim=5),
train_loader = train_loader,
primary_loss_fn = nn.CrossEntropyLoss(),
num_clusters = 4, # K in KMeans
sparsity_weight = 0.01, # α for L₂ sparsity on input weights
cluster_weight = 0.5, # β for cluster‐rep loss
verbose = True # print per-epoch loss summaries
)
trainer.train(
main_epochs = 15, # epochs for joint phase
main_lr = 1e-4, # LR for joint phase
pretrain_epochs = 10, # epochs for pretrain phase
pretrain_lr = 1e-3, # LR for pretrain (defaults to main_lr if None)
train_steps_per_batch = 8, # inner updates per batch in joint phase
)After training, we can then easily extract the important inputs
sorted_idx = trainer.model.get_sorted_input_indices()
print("Top 5 features by importance:", sorted_idx[:5].tolist())
print(trainer.model.get_input_importance())
>> Top 5 features by importance: [3, 1, 4, 2, 0]
>> tensor([0.7594, 0.8327, 0.8003, 0.9258, 0.8141, 0.0107, 0.0199, 0.0329, 0.0043,
0.0025, 0.0448, 0.0054, 0.0119, 0.0021, 0.0190, 0.0055, 0.0063, 0.0073,
0.0059, 0.0189], grad_fn=) Which is awesome, we got back the 5 important inputs as expected!
We can also easily extract the clusters using the representation layer and plot them:
centroids, labels = trainer.get_clusters(dataset)
plt.figure(figsize=(8, 6))
plt.scatter(
components[:, 0],
components[:, 1],
c=labels,
cmap='tab10',
s=20,
alpha=0.7
)
plt.xlabel('Principal Component 1')
plt.ylabel('Principal Component 2')
plt.title('PCA of Synthetic Dataset')
plt.colorbar(label='True Cluster')
plt.tight_layout()
plt.show()
And boom, that’s all!
Conclusion
Though DeepType won’t be the right tool for every problem, it offers a powerful way to integrate domain knowledge into the clustering process. So if you find yourself with a meaningful loss function and a desire to uncover structure in your data—give DeepType a shot!
Please contact mchak@calpoly.edu for any inquiries. All images by author unless stated otherwise.
- Biologists have determined a set of cancer subtypes for the broader category breast cancer. Though I’m no expert, it’s safe to assume that these subtypes were identified by biologists for a reason. The the authors trained their model to predict the subtype for a patient, which provided the biological context necessary to produce novel, interesting clusters. Given the goal, though, I’m not sure why the authors chose to predict on subtypes instead of patient outcomes directly, though — in fact, I bet the results from such an experiment would be interesting.
- The norm presented is defined as

We transpose w since we want to penalize the columns of the weight matrix rather than the rows. This is important because in a fully connected neural network layer, each column of the weight matrix corresponds to an input feature. By applying the ℓ2,1 norm to the transposed matrix, we encourage entire input features to be zeroed out, promoting feature-level sparsity
Cover image source: here
The post Want Better Clusters? Try DeepType appeared first on Towards Data Science.
































































![Apple Developing AI 'Vibe-Coding' Assistant for Xcode With Anthropic [Report]](https://www.iclarified.com/images/news/97200/97200/97200-640.jpg)
![Apple's New Ads Spotlight Apple Watch for Kids [Video]](https://www.iclarified.com/images/news/97197/97197/97197-640.jpg)






































































































_Inge_Johnsson-Alamy.jpg?width=1280&auto=webp&quality=80&disable=upscale#)



















































































































![[The AI Show Episode 145]: OpenAI Releases o3 and o4-mini, AI Is Causing “Quiet Layoffs,” Executive Order on Youth AI Education & GPT-4o’s Controversial Update](https://www.marketingaiinstitute.com/hubfs/ep%20145%20cover.png)































































































































![From Art School Drop-out to Microsoft Engineer with Shashi Lo [Podcast #170]](https://cdn.hashnode.com/res/hashnode/image/upload/v1746203291209/439bf16b-c820-4fe8-b69e-94d80533b2df.png?#)