Lyra: A Computationally Efficient Subquadratic Architecture for Biological Sequence Modeling
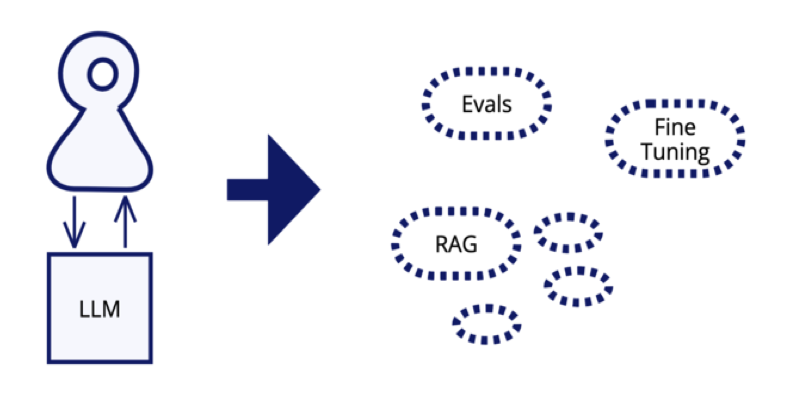
Deep learning architectures like CNNs and Transformers have significantly advanced biological sequence modeling by capturing local and long-range dependencies. However, their application in biological contexts is constrained by high computational demands and the need for large datasets. CNNs efficiently detect local sequence patterns with subquadratic scaling, whereas Transformers leverage self-attention to model global interactions but […] The post Lyra: A Computationally Efficient Subquadratic Architecture for Biological Sequence Modeling appeared first on MarkTechPost.

Deep learning architectures like CNNs and Transformers have significantly advanced biological sequence modeling by capturing local and long-range dependencies. However, their application in biological contexts is constrained by high computational demands and the need for large datasets. CNNs efficiently detect local sequence patterns with subquadratic scaling, whereas Transformers leverage self-attention to model global interactions but require quadratic scaling, making them computationally expensive. Hybrid models, such as Enformers, integrate CNNs and Transformers to balance local and international context modeling, but they still face scalability issues. Large-scale Transformer-based models, including AlphaFold2 and ESM3, have achieved breakthroughs in protein structure prediction and sequence-function modeling. Yet, their reliance on extensive parameter scaling limits their efficiency in biological systems where data availability is often restricted. This highlights the need for more computationally efficient approaches to model sequence-to-function relationships accurately.
To overcome these challenges, epistasis—the interaction between mutations within a sequence—provides a structured mathematical framework for biological sequence modeling. Multilinear polynomials can represent these interactions, offering a principled way to understand sequence-function relationships. State space models (SSMs) naturally align with this polynomial structure, using hidden dimensions to approximate epistatic effects. Unlike Transformers, SSMs utilize Fast Fourier Transform (FFT) convolutions to model global dependencies efficiently while maintaining subquadratic scaling. Additionally, integrating gated depthwise convolutions enhances local feature extraction and expressivity through adaptive feature selection. This hybrid approach balances computational efficiency with interpretability, making it a promising alternative to Transformer-based architectures for biological sequence modeling.
Researchers from institutions, including MIT, Harvard, and Carnegie Mellon, introduce Lyra, a subquadratic sequence modeling architecture designed for biological applications. Lyra integrates SSMs to capture long-range dependencies with projected gated convolutions for local feature extraction, enabling efficient O(N log N) scaling. It effectively models epistatic interactions and achieves state-of-the-art performance across over 100 biological tasks, including protein fitness prediction, RNA function analysis, and CRISPR guide design. Lyra operates with significantly fewer parameters—up to 120,000 times smaller than existing models—while being 64.18 times faster in inference, democratizing access to advanced biological sequence modeling.
Lyra consists of two key components: Projected Gated Convolution (PGC) blocks and a state-space layer with depthwise convolution (S4D). With approximately 55,000 parameters, the model includes two PGC blocks for capturing local dependencies, followed by an S4D layer for modeling long-range interactions. PGC processes input sequences by projecting them to intermediate dimensions, applying depthwise 1D convolutions and linear projections, and recombining features through element-wise multiplication. S4D leverages diagonal state-space models to compute convolution kernels using matrices A, B, and C, efficiently capturing sequence-wide dependencies through weighted exponential terms and enhancing Lyra’s ability to model biological data effectively.
Lyra is a sequence modeling architecture designed to capture local and long-range dependencies in biological sequences efficiently. It integrates PGCs for localized modeling and diagonalized S4D for global interactions. Lyra approximates complex epistatic interactions using polynomial expressivity, outperforming Transformer-based models in tasks like protein fitness landscape prediction and deep mutational scanning. It achieves state-of-the-art accuracy across various protein and nucleic acid modeling applications, including disorder prediction, mutation impact analysis, and RNA-dependent RNA polymerase detection, while maintaining a significantly smaller parameter count and lower computational cost than existing large-scale models.
In conclusion, Lyra introduces a subquadratic architecture for biological sequence modeling, leveraging SSMs to approximate multilinear polynomial functions efficiently. This enables superior modeling of epistatic interactions while significantly reducing computational demands. By integrating PGCs for local feature extraction, Lyra achieves state-of-the-art performance across over 100 biological tasks, including protein fitness prediction, RNA analysis, and CRISPR guide design. It outperforms large foundation models with far fewer parameters and faster inference, requiring only one or two GPUs for training within hours. Lyra’s efficiency democratizes access to advanced biological modeling with therapeutics, pathogen surveillance, and biomanufacturing applications.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, feel free to follow us on Twitter and don’t forget to join our 85k+ ML SubReddit.
The post Lyra: A Computationally Efficient Subquadratic Architecture for Biological Sequence Modeling appeared first on MarkTechPost.




























































![Apple to Avoid EU Fine Over Browser Choice Screen [Report]](https://www.iclarified.com/images/news/96813/96813/96813-640.jpg)




















![iOS 18.4 Release Candidate comes bundled with over 50 new changes and features [Video]](https://i0.wp.com/9to5mac.com/wp-content/uploads/sites/6/2025/03/iOS-18.4-Release-Candidate-Visual-Intelligence-Action-Button-iPhone-16e.jpg?resize=1200%2C628&quality=82&strip=all&ssl=1)







-xl-xl.jpg)













































































_alon_harel_Alamy.jpg?#)





























































![[The AI Show Episode 140]: New AGI Warnings, OpenAI Suggests Government Policy, Sam Altman Teases Creative Writing Model, Claude Web Search & Apple’s AI Woes](https://www.marketingaiinstitute.com/hubfs/ep%20140%20cover.png)
![[The AI Show Episode 139]: The Government Knows AGI Is Coming, Superintelligence Strategy, OpenAI’s $20,000 Per Month Agents & Top 100 Gen AI Apps](https://www.marketingaiinstitute.com/hubfs/ep%20139%20cover-2.png)

























































































































.png?width=1920&height=1920&fit=bounds&quality=80&format=jpg&auto=webp#)
.png?width=1920&height=1920&fit=bounds&quality=80&format=jpg&auto=webp#)